- API identification products are test kits for identification of Gram positive and Gram negative bacteria and yeast.
- API strips give accurate identifications based on extensive databases and are standardized, easy-to-use test systems.
- The kits include strips that contain up to 20 miniature biochemical tests which are all quick, safe and easy to perform.
- API (Analytical Profile Index) 20E is a biochemical panel for identification and differentiation of members of the family Enterobacteriaceae.
- It is hence a well-established method for manual microorganism identification to the species level.
Objective
To identify and differentiate members of family Enterobacteriaceae.
Principle
The API range provides a standardized, miniaturized version of existing identification techniques, which up until now were complicated to perform and difficult to read. In the API 20E, the plastic strip holds twenty mini-test chambers containing dehydrated media having chemically-defined compositions for each test. They usually detect enzymatic activity, mostly related to fermentation of carbohydrate or catabolism of proteins or amino acids by the inoculated organisms.
A bacterial suspension is used to rehydrate each of the wells and the strips are incubated. During incubation, metabolism produces color changes that are either spontaneous or revealed by the addition of reagents. All positive and negative test results are compiled to obtain a profile number, which is then compared with profile numbers in a commercial codebook (or online) to determine the identification of the bacterial species.
The Test Kit
The test kit enables the following tests:
- ONPG: test for β-galactosidase enzyme by hydrolysis of the substrate o-nitrophenyl-b-D-galactopyranoside
- ADH: decarboxylation of the amino acid arginine by arginine dihydrolase
- LDC: decarboxylation of the amino acid lysine by lysine decarboxylase
- ODC: decarboxylation of the amino acid ornithine by ornithine decarboxylase
- CIT: utilization of citrate as only carbon source
- H2S: production of hydrogen sulfide
- URE: test for the enzyme urease
- TDA (Tryptophan deaminase): detection of the enzyme tryptophan deaminase: Reagent- Ferric Chloride.
- IND: Indole Test-production of indole from tryptophan by the enzyme tryptophanase . Reagent- Indole is detected by addition of Kovac’s reagent.
- VP: the Voges-Proskauer test for the detection of acetoin (acetyl methylcarbinol) produced by fermentation of glucose by bacteria utilizing the butylene glycol pathway
- GEL: test for the production of the enzyme gelatinase which liquefies gelatin
- GLU: fermentation of glucose (hexose sugar)
- MAN: fermentation of mannose (hexose sugar)
- INO: fermentation of inositol (cyclic polyalcohol)
- SOR: fermentation of sorbitol (alcohol sugar)
- RHA: fermentation of rhamnose (methyl pentose sugar)
- SAC: fermentation of sucrose (disaccharide)
- MEL: fermentation of melibiose (disaccharide)
- AMY: fermentation of amygdalin (glycoside)
- ARA: fermentation of arabinose (pentose sugar)
Method
- Confirm the culture is of an Enterobacteriaceae. To test this, a quick oxidase test for cytochrome c oxidase may be performed.
- Pick a single isolated colony (from a pure culture) and make a suspension of it in sterile distilled water.
- Take the API20E Biochemical Test Strip which contains dehydrated bacterial media/bio-chemical reagents in 20 separate compartments.
- Using a pasteur pipette, fill up (up to the brim) the compartments with the bacterial suspension.
- Add sterile oil into the ADH, LDC, ODC, H2S and URE compartments.
- Put some drops of water in the tray and put the API Test strip and close the tray.
- Mark the tray with identification number (Patient ID or Organism ID), date and your initials.
- Incubate the tray at 37oC for 18 to 24 hours.
Result Interpretation
- For some of the compartments, the color change can be read straightway after 24 hours but for some reagents must be added to them before interpretation.
- Add following reagents to these specific compartments:
- TDA: Put one drop of Ferric Chloride
- IND: Put one drop of Kovacs reagent
- VP: Put one drop of 40 % KOH (VP reagent 1) & One drop of VP Reagent 2 (α-Naphthol)
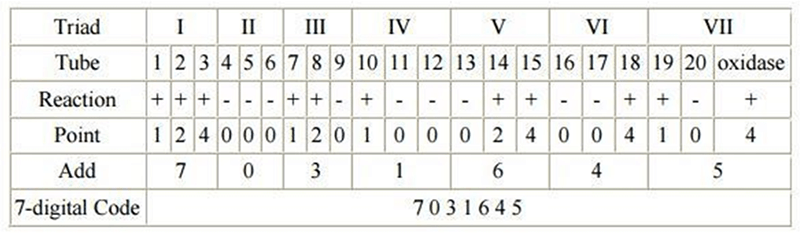
- Get the API Reading Scale (color chart) by marking each test as positive or negative on the lid of the tray. The wells are marked off into triplets by black triangles, for which scores are allocated.

- Add up the scores for the positive wells only in each triplet.
- Three test reactions are added together at a time to give a 7-digit number, which can then be looked up in the codebook. The highest score possible for a triplet is 7 (the sum of 1, 2 and 4) and the lowest is 0.
- Identify the organism by using API catalog or apiweb (online)
Uses
API offers manual identification of microorganisms for:
- Infectious disease diagnosis
- Identification of important industrial microorganisms
Merits
- Fast and efficient (18-24 hour identification of Enterobacteriaceae and other non-fastidious gram negative bacteria)
- Economical to run
- User-friendly
- Standardized
- API strips have a long shelf life, enabling every laboratory to keep the test kits on hand.
- Allow accurate identifications based on extensive databases
References
- https://www.biomerieux-usa.com/clinical/api
- https://bio.libretexts.org/Demos%2C_Techniques%2C_and_Experiments/Microbiology_Labs_I/43%3A_API-20E_multitest_strip
- https://www.jlindquist.com/generalmicro/102bactid2.html
- https://jcm.asm.org/content/jcm/7/6/539.full.pdf
Similar Posts:
- Oxidase Test- Principle, Uses, Procedure, Types, Result Interpretation, Examples and Limitations
- Indole Test- Principle, Reagents, Procedure, Result Interpretation and Limitations
- The Enterotube™ II – Procedure, Result Interpretation, Merits and Limitations
- PYR Test- Principle, Uses, Procedure and Result Interpretation


What are the limitation of Api 20
Is API 20E suitable for other bacteria apart from the enterobacteriaceae
Why citrate is full in api 20e
Question: Which test results can be used to identify Pseudomonas aeruginosa or Acinetobacter baumanii?