The oxidase test detects the presence of a cytochrome oxidase system that will catalyse the transport of electrons between electron donors in the bacteria and a redox dye- tetramethyl-p-phenylene-diamine. The dye is reduced to deep purple color. This test is used to assist in the identification of Pseudomonas, Neisseria, Alcaligens, Aeromonas, Campylobacter, Vibrio, Brucella and Pasteurella, all of which produce the enzyme cytochrome oxidase.
A number of reagents can be used for this test.
- Kovacs Oxidase Reagent:
1% tetra-methyl-p-phenylenediamine dihydrochloride, in water
- Gordon and McLeod’s Reagent:
1% dimethyl-p-phenylenediamine dihydrochloride, in water
- Gaby and Hadley (indophenol oxidase) Reagent:
1% α-naphthol in 95% ethanol
1% p-aminodimethylaniline HCL
Principle of Oxidase Test
Cytochrome containing organisms produce an intracellular oxidase enzyme. This oxidase enzyme catalyzes the oxidation of cytochrome c. Organisms which contain cytochrome c as part of their respiratory chain are oxidase-positive and turn the reagent blue/purple. Organisms lacking cytochrome c as part of their respiratory chain do not oxidize the reagent, leaving it colorless within the limits of the test, and are oxidase-negative.
Oxidase positive bacteria possess cytochrome oxidase or indophenol oxidase (an iron containing haemoprotein). Both of these catalyse the transport of electrons from donor compounds (NADH) to electron acceptors (usually oxygen). The test reagent, N, N, N’, N’-tetramethyl-p-phenylenediamine dihydrochloride acts as an artificial electron acceptor for the enzyme oxidase. The oxidised reagent forms the colored compound indophenol blue.
The cytochrome system is usually only present in aerobic organisms which are capable of utilising oxygen as the final hydrogen receptor. The end product of this metabolism is either water or hydrogen peroxide (broken down by catalase).
Purpose/Uses of Oxidase Test
- The oxidase test is used to determine if an organism possesses the cytochrome oxidase enzyme.
- The test is used as an aid for the differentiation of Neisseria, Moraxella, Campylobacter and Pasteurella species (oxidase positive).
- It is also used to differentiate pseudomonads from related species.
Procedure of Oxidase Test
There are many method variations to the oxidase test. These include, but are not limited to, the filter paper test, direct plate method, swab method, impregnated oxidase test strip method and test tube method. All times and concentrations are based upon the original recommendations.
Dry Filter Paper Method

Since the oxidase reagent is unstable and has to be freshly prepared for use, this method is convenient.
- Strip of Whatman’s No. 1 filter paper are soaked in a freshly prepared 1% solution of tertramethyl-p-phenylene-diamine dihydrochloride.
- After draining for about 30 seconds, the strips are freeze dried and stored in a dark bottle tightly sealed with a screw cap.
- For use, a strip is removed, laid in a petri dish and moistened with distilled water.
- The colony to be tested is picked up with a platinum loop and smeared over the moist area.
- A positive reaction is indicated by an intense deep-purple hue, appearing within 5-10 seconds, a “delayed positive” reaction by coloration in 10-60 seconds, and a negative reaction by absence of coloration or by coloration later than 60 seconds.
Wet Filter Paper Method
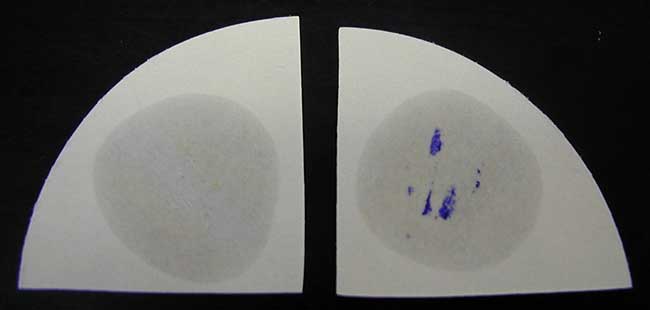
- A strip of filter paper is soaked with a little freshly made 1% solution of the reagent.
- A speck of culture is rubbed on it with a platinum loop.
- A positive reaction is indicated by an intense deep-purple hue, appearing within 5-10 seconds, a “delayed positive” reaction by coloration in 10-60 seconds, and a negative reaction by absence of coloration or by coloration later than 60 seconds.
Direct Plate Method
- Add 2 -3 drops of reagent directly to suspect colonies on an agar plate. Do not flood the plate with the reagent.
- Observe for color change within 10 seconds.
Note: The Direct Plate method should be carried out on a non-selective agar plate.
- When using Kovac’s oxidase reagent, microorganisms are oxidase positive when the color changes to dark purple within 5 to 10 seconds. Microorganisms are delayed oxidase positive when the color changes to purple within 60 to 90 seconds. Microorganisms are oxidase negative if the color does not change or it takes longer than 2 minutes.
- When using Gordon and McLeod reagent, microorganisms are oxidase positive when the color changes to red within 10 to 30 minutes or to black within 60 minutes. Microorganisms are oxidase negative if the color does not change.
Swab Method
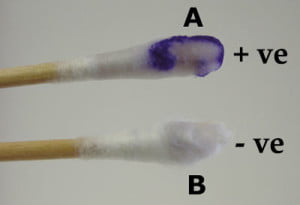
- Dip swab into reagent and then touch an isolated suspect colony
- Observe for color change within 10 seconds.
Test Tube Method
- Grow a fresh culture (18 to 24 hours) of bacteria in 4.5 ml of nutrient broth (or standard media that does not contain a high concentration of sugar).
- Add 0.2 ml of 1% α-naphthol, then add 0.3 ml of 1% p-aminodimethylaniline oxalate (Gaby and Hadley reagents).
- Shake vigorously to ensure mixing and thorough oxygenation of the culture.
- Observe for color changes.
- Microorganisms are oxidase positive when the color changes to blue within 15 to 30 seconds. Microorganisms are delayed oxidase positive when the color changes to purple within 2 to 3 minutes. Microorganisms are oxidase negative if the color does not change.
Result Interpretation of Oxidase Test
Positive Result
Development of a deep purple-blue/blue color indicates oxidase production within 5-10 seconds.
Negative Result
No purple-blue color/No color change.
Examples
Oxidase Positive Organisms: Pseudomonas, Neisseria, Alcaligens, Aeromonas, Campylobacter, Vibrio, Brucella, Pasteurella, Moraxella, Helicobacter pylori, Legionella pneumophila, etc.
Oxidase Negative Organisms: Enterobacteriaceae (e.g. E. coli)
Quality Control for Oxidase Test
Positive Control: Pseudomonas aeruginosa ATCC 27853
Negative Control: Escherichia coli ATCC 25922
Limitations of Oxidase Test
- The reagents used in the oxidase test have been shown to auto-oxidize, so it is very important to use fresh reagents, no older than 1 week.
- Both bacteria and yeast grown on media containing high concentrations of glucose show inhibited oxidase activity, so it is recommended to test colonies grown on media without excess sugar, such as nutrient agar. Tryptic soy agar is also an excellent media.
- Bacteria grown on media containing dyes may give aberrant results.
- The test reagents will effectively kill the microorganisms, so sub-culturing should be done prior to adding any reagent to an active culture.
- The oxidase test can be used in the presumptive identification of Neisseria and in the differentiation and identification of gram-negative bacilli. Oxidase-positive organisms should be examined by gram stain to determine morphology and gram reaction. Additional biochemical tests are recommended for complete identification.
- Use of a nichrome or other iron containing loop may yield false-positive reactions. Platinum loops are recommended.
- Most Haemophilus are oxidase-positive. Less sensitive strips or reagents may yield false-negative results.
- Oxidase reactions of gram-negative bacilli should be determined on non-selective and non-differential media to ensure valid results. Also, colonies taken from media containing high levels of glucose may give false-negative reactions.
- It is recommended to use colonies that are 18-24 hours old. Older colonies will produce weaker reactions.
- Any color changes appearing after 20 seconds should be disregarded.

please guide
I have found OXIDASE POSITIVE result from the colonies grown on MITIS SALIVARIUS AGAR (having bacitracin or potassium tellurite) which is the selective media for STREPTOCOCCUS MUTANS.
How is this possible as that s.mutans appear oxidase negative.
What is going wrong i couldn’t understand.
You perform the oxidase test on your unknown bacterium and it turns blue after 2 minutes. Does this mean a positive result? Explain.
yes it means its a +ve result.
hi ,
i think Haemophilus is regarded as a “late oxydase positive” organism i.e positive 30 to 60 seconds (as opposed to immediate positve organisms)
usually we use Escherichia coli ATCC as a negative control on abrerant late positive results
what are your thoughts on this ?
thank you
Hello please is Borrelia a positive oxidase or negative?
Please reply me back as soon as possible.
Thanks
Borrelia is oxidase positive
i have tetra-methyl-p-phenylenediamine dihydrochloride, powder 1 gram i just i want to know how much ml i mix with mater?
Please help me..
Thanks
@Jonnel Penascosa you need 99 mL water, preferably sterile, but normal distilled would work also.
If I’m not wrong, that must be 100mL because the one we are mixing is a solid and not a liquid, so to prepare 1% stock 1gm of the powder should be dissolved in 100mL sterile distilled water.
100ml distilled water
I’ve appreciated modifications/suggestions to improve the TMPD test.
However, though this simple test is known since long time, it is ambiguous in the interpretation of results and gives wrong information required in bacterial identification.
It is not sensitive to detect cytochrome oxidase(s) activity in Enterobacteriaceae and other aerobic bacteria, which obviously contain corresponding gene(s). Therefore, Escherichia coli can’t be considered as a negative control; the test simply does not work on these cells!
The problem might be related to TMPD transfer through cell membrane; positive reactions were described with extracts of “negative controls” using TMPD.
No, it means the bacteria was an oxidase negative. When it takes more than 2 minutes to colour change, it may the reason of some contaminated microbes from the air between the 2 minutes
Escherichia coli is a facultative anaerobe, not obligate aerobe so probably has different genes in its electron transport chain. The oxidase test is meant to detect cytochrome oxidase which is found in obligate aerobes. Since E.coli is a facultative anaerobe it is oxidase negative.
Hello,
Could we perform the oxidase test from colonies taken directly from selective media such as Chromocult Coliform agar, CCA? ( I’m using oxidase strip )
Tq
Zam
Microbiologist